Models and Simulations | Ion Channels | Cardiac Cells | Cardiac Tissue
We develop mathematical Markov models of ionic currents. Such models reflect the stochastic, memory-less processes which govern ion channel behavior. In the past, we have developed models of INa, ICa(L), IKr, and IKs. We have used these models to draw general conclusions about ion channel behavior, with our major emphasis placed on the role of channel kinetics in the ventricular action potential. In several studies, we build models for wild type and mutant channels which are then placed in the context of the action potential where arrhthmogenic features of the mutation can be identified and addressed.
Recently, our work has focused on using crystal structure homology models to relate channel biochemistry and biophysics to voltage dependent gating. We use molecular dynamics to simulate the ion-channel protein. This method allows us to build a model for channel current that comes directly from protein structure.
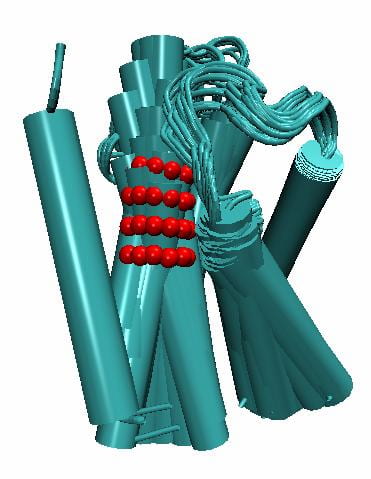
Starting from a deep closed state at -100mV, the membrane potential is raised to +50mV. Motion of one voltage sensor segment(S3-S4 complex) is simulated under the influence of a stochastic force field. The presented motion trajectory is one of infinite possibilities. The four S6 segments of the tetrameric channel are assumed symmetric (only two are shown, S6 and S6′). (A. Nekouzadeh, Y. Rudy, 2015)
To relate KCNQ1 and IKs dynamics and molecular interactions to the channel and cardiac-cell electrophysiology, we will combine molecular dynamics simulation methodology and algorithms with models of ion-channel kinetics and of the whole-cell action potential. For the molecular (structural) dynamics simulations, key experimental data come from the recently published crystal structure of Kv1.2 (a Shaker homologue in rat), which provide the locations of conserved charged amino acids in the channel voltage sensor . By combining this information with the wealth of electrophysiological recordings of Shaker , KCNQ1, and IKs currents, we will construct a homologous model of IKs activation that is based on the electrostatic interaction between the KCNQ1 voltage sensor (S4) and its surrounding channel environment.
For more information, see review articles:
A. Nekouzadeh, Y. Rudy. “Conformational changes of an ion-channel during gating and emerging electrophysiologic properties: Application of a computational approach to cardiac Kv7.1.” Prog Biophys Mol Biol. 2016 Jan;120(1-3):18-27. 2015.12.014. Epub 2015 Dec 30. PMCID: PMC4955398.
Y. Rudy, J.R. Silva, “Computational Biology in the Study of Cardiac Ion Channels and Cell Electrophysiology” Quarterly Reviews of Biophysics 2006; 39:57-116. PMCID: PMC1994938